Note
Go to the end to download the full example code.
Quantile prediction with Random Forest Regressor class#
An example that demonstrates how to use the Random Forest to generate quantile predictions such as conditional median and prediction intervals. The example compares the predictions to a ground truth function used to generate noisy samples.
This example was heavily inspired by quantile-forest package. See their package here.
from collections import defaultdict
import matplotlib.pyplot as plt
import numpy as np
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
Generate the data
def make_toy_dataset(n_samples, seed=0):
rng = np.random.RandomState(seed)
x = rng.uniform(0, 10, size=n_samples)
f = x * np.sin(x)
sigma = 0.25 + x / 10
noise = rng.lognormal(sigma=sigma) - np.exp(sigma**2 / 2)
y = f + noise
return np.atleast_2d(x).T, y
n_samples = 1000
X, y = make_toy_dataset(n_samples)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
xx = np.atleast_2d(np.linspace(0, 10, n_samples)).T
Fit the model to the training samples#
rf = RandomForestRegressor(max_depth=3, random_state=0)
rf.fit(X_train, y_train)
y_pred = rf.predict(xx)
# get the leaf nodes that each sample fell into
leaf_ids = rf.apply(X_train)
# create a list of dictionary that maps node to samples that fell into it
# for each tree
node_to_indices = []
for tree in range(leaf_ids.shape[1]):
d = defaultdict(list)
for id, leaf in enumerate(leaf_ids[:, tree]):
d[leaf].append(id)
node_to_indices.append(d)
# drop the X_test to the trained tree and
# get the indices of leaf nodes that fall into it
leaf_ids_test = rf.apply(xx)
# for each samples, collect the indices of the samples that fell into
# the same leaf node for each tree
y_pred_quatile = []
for sample in range(leaf_ids_test.shape[0]):
li = [
node_to_indices[tree][leaf_ids_test[sample][tree]] for tree in range(leaf_ids_test.shape[1])
]
# merge the list of indices into one
idx = [item for sublist in li for item in sublist]
# get the y_train for each corresponding id
y_pred_quatile.append(y_train[idx])
# get the quatile preditions for each predicted sample
y_pred_low = [np.quantile(y_pred_quatile[i], 0.025) for i in range(len(y_pred_quatile))]
y_pred_med = [np.quantile(y_pred_quatile[i], 0.5) for i in range(len(y_pred_quatile))]
y_pred_upp = [np.quantile(y_pred_quatile[i], 0.975) for i in range(len(y_pred_quatile))]
Plot the results#
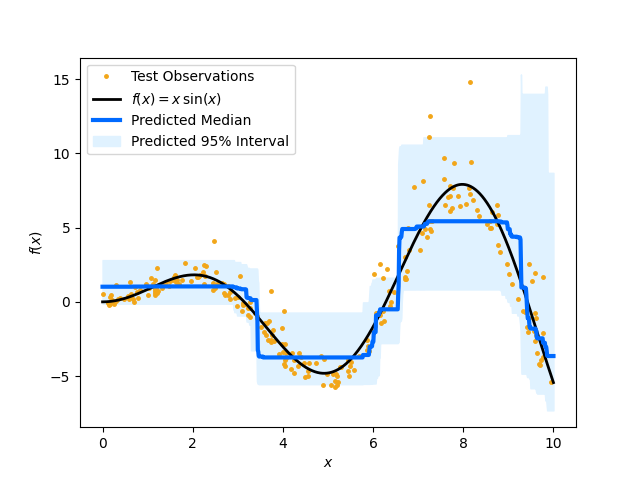
Plot the conditional median and prediction intervals. The blue line is the predicted median and the shaded area indicates the 95% confidence interval of the prediction. The dots are the training data and the black line indicates the function that is used to generated those samples.
plt.plot(X_test, y_test, ".", c="#f2a619", label="Test Observations", ms=5)
plt.plot(xx, (xx * np.sin(xx)), c="black", label="$f(x) = x\,\sin(x)$", lw=2)
plt.plot(xx, y_pred_med, c="#006aff", label="Predicted Median", lw=3, ms=5)
plt.fill_between(
xx.ravel(),
y_pred_low,
y_pred_upp,
color="#e0f2ff",
label="Predicted 95% Interval",
)
plt.xlabel("$x$")
plt.ylabel("$f(x)$")
plt.legend(loc="upper left")
plt.show()
Total running time of the script: (0 minutes 3.149 seconds)
Estimated memory usage: 320 MB