Multiple Network Models
Contents
4.7. Multiple Network Models#
Up to this point, you have studied network models which are useful for a single network. What do you do if you have multiple networks?
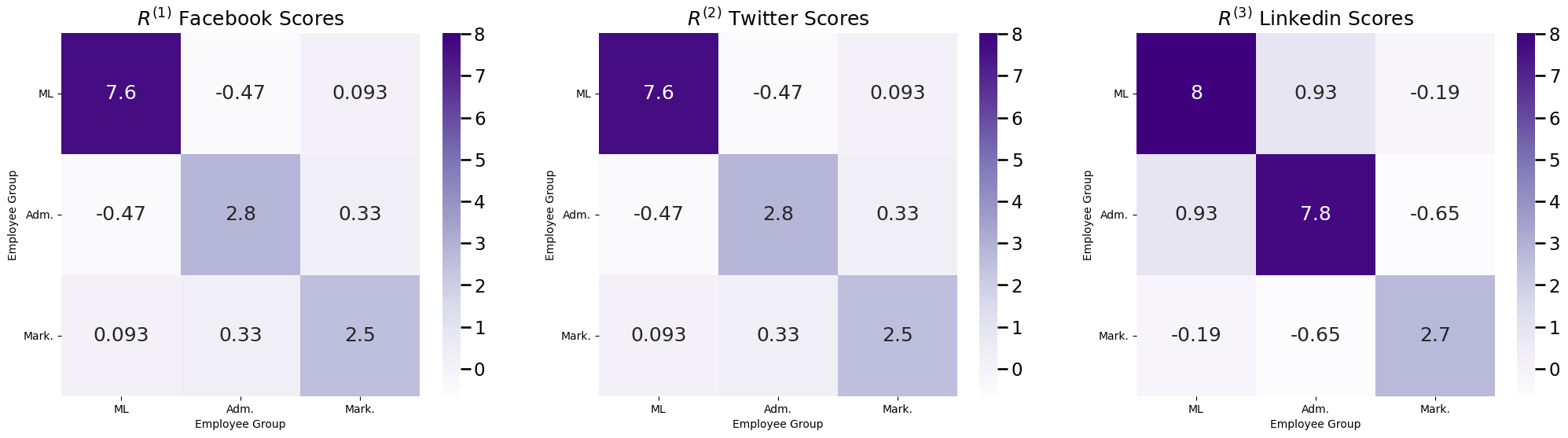
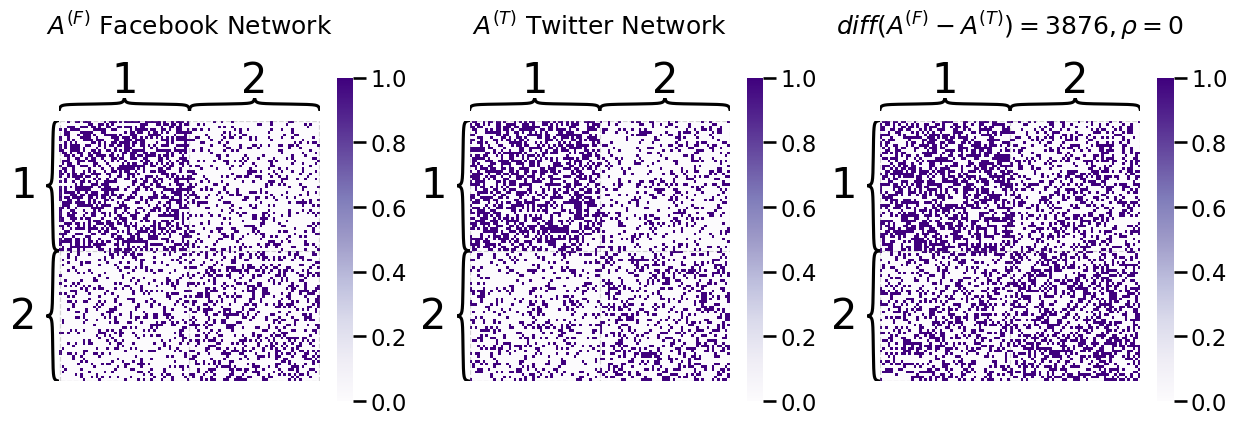
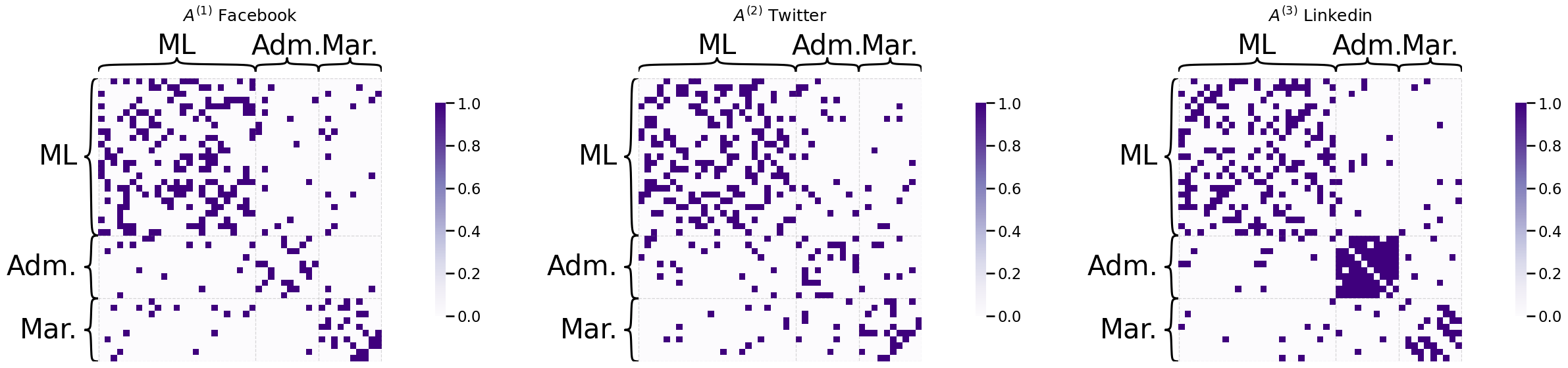
Let’s imagine that you have a company with \(45\) total employees. \(10\) of these employees are company administrative executives, \(25\) of these employees are network machine learning experts, and \(10\) of these employees are marketing experts. You study the social media habits of your employees on Facebook, Twitter, and Linkedin. For a given social networking site, an edge is said to exist between a pair of employees if they are connected on the social media site (by being friends, following one another, or being connected, respectively). As it turns out, individuals tend to most closely associate with the colleagues whom they work most closely: you might expect some sort of community structure to your network, wherein network machine learning experts are more connected with network machine learning experts, marketing experts are more connected with marketing experts, so on and so forth. What you will see below is that all of the networks appear to have the same community organization, though on Linkedin, you see that the administrative executives tend to be a little more connected. This is reflected in the fact that there are more connections between Adm. members on Linkedin:
from graphbook_code import heatmap
from graspologic.simulations import sbm
import numpy as np
import matplotlib.pyplot as plt
K = 3
B1 = np.zeros((K,K))
B1[0,0] = 0.3
B1[0,1] = .05
B1[1,0] = .05
B1[0,2] = .05
B1[2,0] = .05
B1[1,1] = 0.3
B1[1,2] = .05
B1[2,1] = .05
B1[2,2] = 0.3
B2 = B1
B3 = np.copy(B1)
B3[1,1] = 0.9
ns = [25, 10, 10]
A1 = sbm(n=ns, p=B1, directed=False, loops=False)
ys = ["ML" for i in range(0, ns[0])] + ["Adm." for i in range(ns[0],ns[0] + ns[1])] + ["Mar." for i in range(ns[0] + ns[1],ns[0] + ns[1] + ns[2])]
A2 = sbm(n=ns, p=B2, directed=False, loops=False)
A3 = sbm(n=ns, p=B3, directed=False, loops=False)
fig, axs = plt.subplots(1,3, figsize=(30, 6))
heatmap(A1, ax=axs[0], title="$A^{(1)}$ Facebook", color="sequential", xticklabels=False, yticklabels=False, inner_hier_labels=ys);
heatmap(A2, ax=axs[1], title="$A^{(2)}$ Twitter", color="sequential", xticklabels=False, yticklabels=False, inner_hier_labels=ys);
heatmap(A3, ax=axs[2], title="$A^{(3)}$ Linkedin", color="sequential", xticklabels=False, yticklabels=False, inner_hier_labels=ys);
Remember that a random network has an adjacency matrix denoted by a boldfaced uppercase \(\mathbf A\), and has network samples \(A\) (which you just call “networks”). When you have multiple networks, you will need to be able to index them individually. For this reason, in this section, you will use the convention that a random network’s adjacency matrix is denoted by a boldfaced uppercase \(\mathbf A^{(m)}\), where \(m\) tells you which network in your collection you are talking about. The capital letter \(M\) defines the total number of random networks in your collection. In your social network example, since you have connection networks for \(3\) sites, \(N\) is \(3\). When you use the letter \(m\) itself, you will typically be referring to an arbitrary random network among the collection of random networks, where \(m\) is between \(1\) and \(N\). When you have \(M\) total networks, you will write down the entire collection of random networks using the notation \(\left\{\mathbf A^{(1)}, ..., \mathbf A^{(M)}\right\}\). With what you already know, for a random network \(\mathbf A^{(m)}\), you would be able to use a single nework model to describe \(\mathbf A^{(m)}\). This means, for instance, if you thought that each social network could be represented by a different RDPG, that you would have a different latent position matrix \(X^{(m)}\) to define each of the \(30\) networks. In symbols, you would write that each \(\mathbf A^{(m)}\) is an RDPG random nework with latent position matrix \(X^{(m)}\). What is the problem with this description?
What this description lacks is that the three networks share a lot of common information. You might expect that, on some level, the latent position matrices should also show some sort of common structure. However, since you used a unique latent position matrix \(X^{(m)}\) for each random network \(\mathbf A^{(m)}\), you have inherently stated that you think that the networks have completely distinct latent position matrices. If you were to perform a task downstream, such as whether you could identify which employees are in which community, you would have to analyze each latent position matrix individually, and you would not be able to learn a latent position matrix with shared structure across the three networks. Before you jump into multiple network models, let’s provide some context as to how you will build these up.
In the below descriptions, you will tend to build off the Random Dot Product Graph (RDPG), and closely related variations of it. Why? Well, as it turns out, the RDPG is extremely flexible, in that you can represent both ER and SBMs as RDPGs, too. You’ll remember from The IER network unifies independent-edge random networks, as it is the most general independent-edge random network model. What this means is that all models discussed so far, and many not discussed but found in the appendix, can be conceptualized as special cases of the IER network. Stated in other words, a probability matrix is the fundamental unit necessary to describe an independent-edge random network model. For the models covered so far, all GRDPGs are IER networks (by taking P to be XY^\top), all RDPGs are GRDPGs (by taking the right latent positions Y to be equal to the left latent positions X), all SBMs are RDPGs, and all ERs are SBMs, whereas the converses in general are not true. that the ER network was a special case of the SBM and SBMs were special cases of RDPG networks, so the RDPG can describe any network which is an RDPG, an SBM, or an ER random network. This means that building off the RDPG gives you multiple random network models that will be inherently flexible. Further, as you will see in the later section on Learning Network Representations, the RDPG is extremely well-suited for the situation in which you want to analyze SBMs, but do not know which communities the nodes are in ahead of time. This situation is extremely common across numerous disciplines of network machine learning, such as social networking, neuroscience, and many other fields.
So, how can you model your collection of random networks with shared structure?
4.7.1. Joint Random Dot Product Graphs (JRDPG) Model#
In our example on social networks, notice that the Facebook and Twitter connections do not look too qualitatively different. It looks like they have relatively similar connectivity patterns between the different employee working groups, and you might even think that the underlying random network that governs these two social networks are identical. In statisical science, when we have a collection of \(M\) random networks that have the same underlying random process, we describe this with the term homogeneity. Let’s put what this means into context using your coin flip example. If a pair of coins are homogeneous, this means that the probability that they land on heads is identical. Likewise, this intuition extends directly to random networks.
A homogeneous collection of independent-edge random networks \(\left\{\mathbf A^{(1)}, ..., \mathbf A^{(M)}\right\}\) is one in which all of the \(M\) random networks have the same probability matrix, and consequently have the same distribution (statistically, the term is identically distributed). Remember that the probability matrix \(P^{(m)}\) is the matrix whose entries \(p^{(m)}_{ij}\) indicate the probability that an edge exists between nodes \(i\) and \(j\). This is because the probability matrix is the fundamental unit which can be used to describe independent-edge random networks, as we learned in Inhomogeneous Erdos Renyi (IER) Random Network Model. On the other hand, a heterogeneous collection of random networks is a collection of networks \(\left\{\mathbf A^{(1)}, ..., \mathbf A^{(M)}\right\}\) is one in which the probability matrices are not the same for all of the \(M\) networks, and hence, they are not identically distributed.
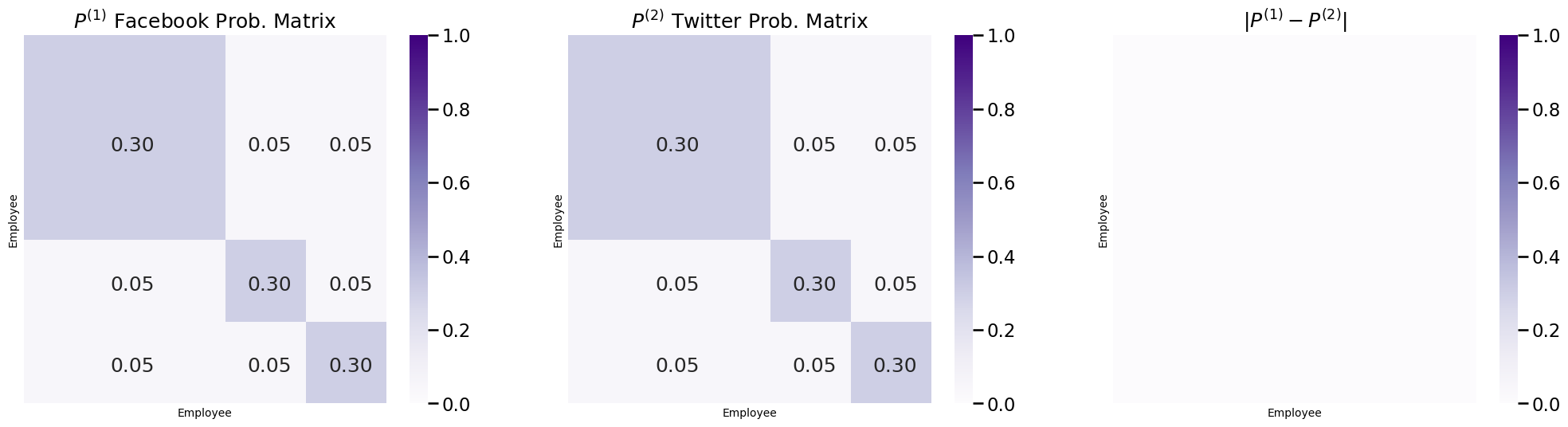
The probability matrices \(P^{(1)}\) and \(P^{(2)}\) for the random networks \(\mathbf A^{(1)}\) and \(\mathbf A^{(2)}\) for Facebook and Twitter from the example we introduced above are shown in the following figure. You can also show the difference between the two probability matrices, to make really clear that they are the same:
import seaborn as sns
from graphbook_code import cmaps
import pandas as pd
def p_from_block(B, ns):
P = np.zeros((np.sum(ns), np.sum(ns)))
ns_cumsum = np.append(0, np.cumsum(ns))
for k, n1 in enumerate(ns_cumsum[0:(len(ns_cumsum)-1)]):
for l, n2 in enumerate(ns_cumsum[k:(len(ns_cumsum)-1)]):
P[ns_cumsum[k]:ns_cumsum[k+1], ns_cumsum[l+k]:ns_cumsum[l+k+1]] = B[k,l+k]
P[ns_cumsum[l+k]:ns_cumsum[l+k+1],ns_cumsum[k]:ns_cumsum[k+1]] = B[l+k,k]
return P
P1 = p_from_block(B1, ns)
U1, S1, V1 = np.linalg.svd(P1)
X1 = U1[:,0:3] @ np.sqrt(np.diag(S1[0:3]))
P2 = p_from_block(B2, ns)
U2, S2, V2 = np.linalg.svd(P2)
X2 = U2[:,0:3] @ np.sqrt(np.diag(S2[0:3]))
P3 = p_from_block(B3, ns)
U3, S3, V3 = np.linalg.svd(P3)
X3 = U3[:,0:3] @ np.sqrt(np.diag(S3[0:3]))
def plot_prob_block_annot(X, title="", nodename="Employee", nodetix=None,
nodelabs=None, ax=None, annot=True):
if (ax is None):
fig, ax = plt.subplots(figsize=(8, 6))
with sns.plotting_context("talk", font_scale=1):
if annot:
X_annot = np.empty(X.shape, dtype='U4')
X_annot[13, 13] = str("{:.3f}".format(X[13,13]))
X_annot[30, 30] = str("{:.3f}".format(X[30,30]))
X_annot[40,40] = str("{:.3f}".format(X[40,40]))
X_annot[13, 30] = str("{:.3f}".format(X[13,30]))
X_annot[30,13] = str("{:.3f}".format(X[30,13]))
X_annot[13,40] = str("{:.3f}".format(X[23,40]))
X_annot[40,13] = str("{:.3f}".format(X[40,13]))
X_annot[30,40] = str("{:.3f}".format(X[30,40]))
X_annot[40,30] = str("{:.3f}".format(X[40,30]))
ax = sns.heatmap(X, cmap="Purples",
ax=ax, cbar_kws=dict(shrink=1), yticklabels=False,
xticklabels=False, vmin=0, vmax=1, annot=pd.DataFrame(X_annot),
fmt='')
else:
ax = sns.heatmap(X, cmap="Purples",
ax=ax, cbar_kws=dict(shrink=1), yticklabels=False,
xticklabels=False, vmin=0, vmax=1)
ax.set_title(title)
cbar = ax.collections[0].colorbar
ax.set(ylabel=nodename, xlabel=nodename)
if (nodetix is not None) and (nodelabs is not None):
ax.set_yticks(nodetix)
ax.set_yticklabels(nodelabs)
ax.set_xticks(nodetix)
ax.set_xticklabels(nodelabs)
cbar.ax.set_frame_on(True)
return
fig, axs = plt.subplots(1,3, figsize=(25, 6))
plot_prob_block_annot(P1, ax=axs[0], title="$P^{(1)}$ Facebook Prob. Matrix")
plot_prob_block_annot(P2, ax=axs[1], title="$P^{(2)}$ Twitter Prob. Matrix")
plot_prob_block_annot(np.abs(P1 - P2), ax=axs[2], annot=False, title="$|P^{(1)} - P^{(2)}|$")
The Joint Random Dot Product Graphs (JRDPG) is the simplest way you can extend the RDPG random network model to multiple random networks. The way you can think of the JRDPG model is that for each of your \(M\) total random neworks, the edges depend on a latent position matrix \(X\). You say that a collection of random networks \(\left\{\mathbf A^{(1)}, ..., \mathbf A^{(N)}\right\}\) with \(n\) nodes is \(JRDPG_{n,M}(X)\) if each random network \(\mathbf A^{(m)}\) is \(RDPG_n(X)\) and if the \(N\) networks are independent. The joint random dot product graph model is formally described by [3], and has many connections with the omnibus embedding [2], which you will learn about in Section 5.4.
4.7.1.1. The JRDPG model does not allow you to convey unique aspects about the networks#
Under the JRDPG model, each of the \(M\) random networks share the same latent position matrix. Remember that for an RDPG, the probability matrix \(P = XX^\top\). This means that for all of the \(M\) networks, \(P^{(m)} = XX^\top\) under the JRDPG model. hence, \(P^{(1)} = P^{(2)} = ... = P^{(M)}\), and all of the probability matrices are identical! This means that the \(M\) random networks are homogeneous. Consequently, the JRDPG can be literally thought of as \(M\) independent RDPGs.
For an RDPG, since \(P = XX^\top\), the probability matrix depends only on the latent positions \(X\). This means that you can tell whether a collection of networks are homogeneous just by looking at the latent position matrices! It turns out that the random networks underlying the samples for the social networks in your given example were just SBMs. From the section on Random Dot Product Graphs (RDPG), you learned that SBMs are just RDPGs with a special latent position matrix. Let’s try this first by looking at latent position matrices for \(\mathbf A^{(1)}\) and \(\mathbf A^{(2)}\) from the random networks for Facebook and Twitter first which were used to generate our network samples:
from numpy.linalg import svd
def plot_latent(X, title="", nodename="Employee", ylabel="Latent Dimension",
nodetix=None, vmin=None, vmax=None, nodelabs=None, ax=None, dimtix=None, dimlabs=None):
if ax is None:
fig, ax = plt.subplots(figsize=(6, 6))
with sns.plotting_context("talk", font_scale=1):
lim_max = np.max(np.abs(X))
if vmin is None:
vmin = -lim_max
if vmax is None:
vmax = lim_max
X_annot = np.empty((45, 3), dtype='U6')
X_annot[13,0] = str("{:.3f}".format(X[13,0]))
X_annot[30,0] = str("{:.3f}".format(X[30,0]))
X_annot[40,0] = str("{:.3f}".format(X[40,0]))
X_annot[13,1] = str("{:.3f}".format(X[13,1]))
X_annot[30,1] = str("{:.3f}".format(X[30,1]))
X_annot[40,1] = str("{:.3f}".format(X[40,1]))
X_annot[13,2] = str("{:.3f}".format(X[13,2]))
X_annot[30,2] = str("{:.3f}".format(X[30,2]))
X_annot[40,2] = str("{:.3f}".format(X[40,2]))
ax = sns.heatmap(X, cmap=cmaps["divergent"],
ax=ax, cbar_kws=dict(shrink=1), yticklabels=False,
xticklabels=False, vmin=vmin, vmax=vmax,
annot=X_annot, fmt='')
ax.set_title(title)
cbar = ax.collections[0].colorbar
ax.set(ylabel=nodename, xlabel=ylabel)
if (nodetix is not None) and (nodelabs is not None):
ax.set_yticks(nodetix)
ax.set_yticklabels(nodelabs)
if (dimtix is not None) and (dimlabs is not None):
ax.set_xticks(dimtix)
ax.set_xticklabels(dimlabs)
cbar.ax.set_frame_on(True)
return
fig, axs = plt.subplots(1,3, figsize=(20, 6))
vmin = np.min([X1,X2,X3]); vmax = np.max([X1, X2, X3])
plot_latent(X1, nodetix=[0,24,34,44], vmin=vmin, vmax=vmax, nodelabs=["1", "25", "35", "45"],
dimtix=[0.5,1.5,2.5], ax=axs[0], dimlabs=["1", "2", "3"], title="$X^{(1)}$ Facebook LPM")
plot_latent(X2, nodetix=[0,24,34,44], nodelabs=["1", "25", "35", "45"],
dimtix=[0.5,1.5,2.5], ax=axs[1], vmin=vmin, vmax=vmax, dimlabs=["1", "2", "3"], title="$X^{(2)}$ Twitter LPM")
plot_latent(np.abs(X1 - X2), nodetix=[0,24,34,44], vmin=vmin, vmax=vmax, nodelabs=["1", "25", "35", "45"],
dimtix=[0.5,1.5,2.5], ax=axs[2], dimlabs=["1", "2", "3"], title="$|X^{(1)} - X^{(2)}|$")
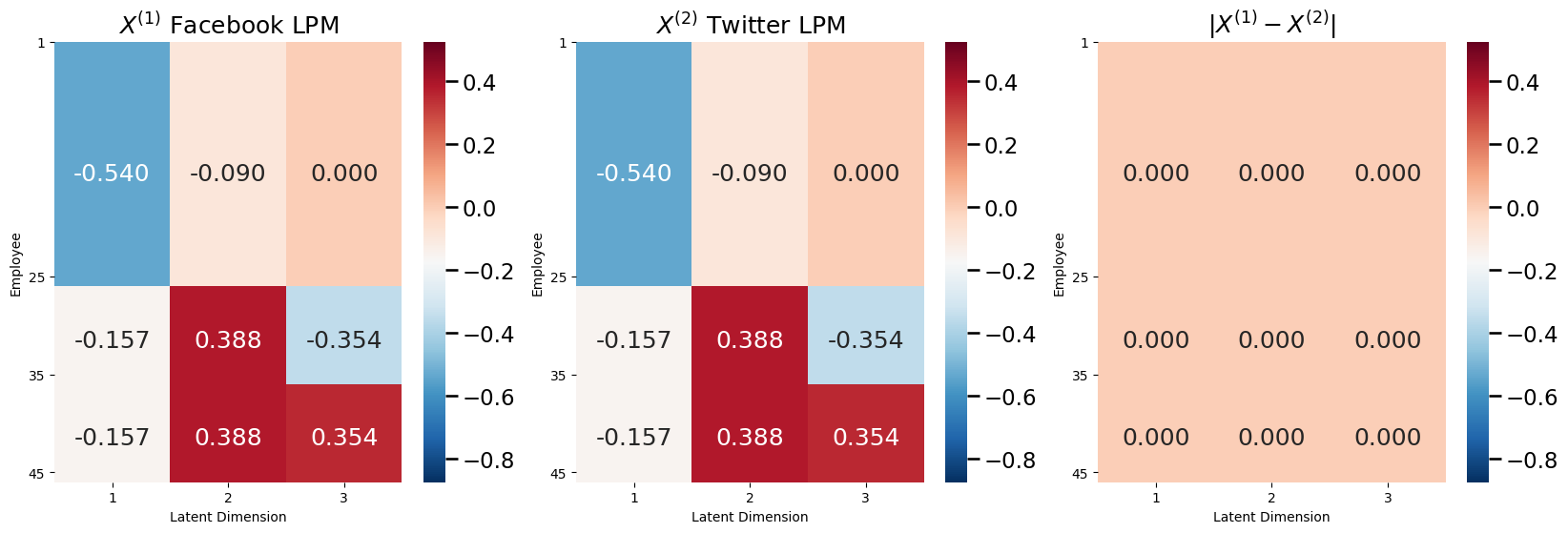
So the latent position matrices for Facebook and Twitter are exactly identical. Therefore, the collection of random networks \(\left\{\mathbf A^{(1)}, \mathbf A^{(2)}\right\}\) are homogeneous, and you could model this pair of networks using the JRDPG.
What about Linkedin? Well, as it turns out, Linkedin had a different latent position matrix from both Facebook and Twitter:
fig, axs = plt.subplots(1,3, figsize=(20, 6))
vmin = np.min([X1,X2,X3]); vmax = np.max([X1, X2, X3])
plot_latent(X1, nodetix=[0,24,34,44], vmin=vmin, vmax=vmax, nodelabs=["1", "25", "35", "45"],
dimtix=[0.5,1.5,2.5], ax=axs[0], dimlabs=["1", "2", "3"], title="$X^{(1)}$ Facebook LPM")
plot_latent(X3, nodetix=[0,24,34,44], nodelabs=["1", "25", "35", "45"],
dimtix=[0.5,1.5,2.5], ax=axs[1], vmin=vmin, vmax=vmax, dimlabs=["1", "2", "3"], title="$X^{(3)}$ Linkedin LPM")
plot_latent(np.abs(X1 - X3), nodetix=[0,24,34,44], vmin=vmin, vmax=vmax, nodelabs=["1", "25", "35", "45"],
dimtix=[0.5,1.5,2.5], ax=axs[2], dimlabs=["1", "2", "3"], title="$|X^{(1)} - X^{(3)}|$")
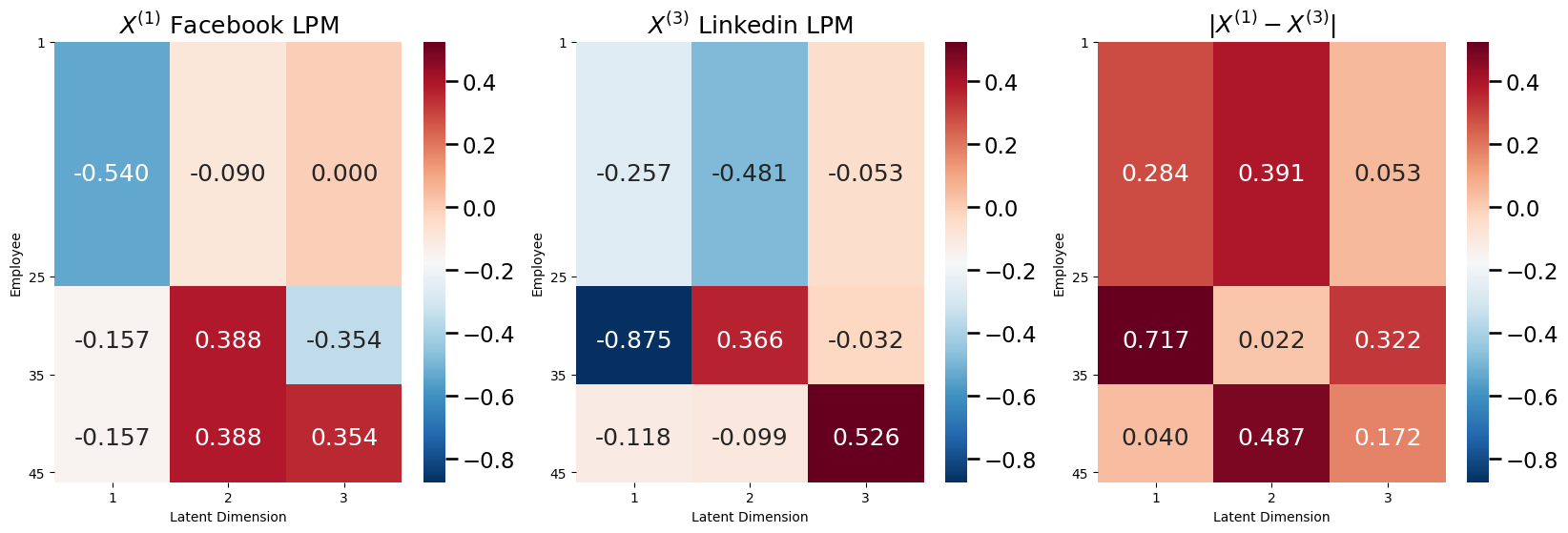
So, \(\mathbf A^{(1)}\) and \(\mathbf A^{(3)}\) do not have the same latent position matrices. This means that the collections of random networks \(\left\{\mathbf A^{(1)}, \mathbf A^{(3)}\right\}\), \(\left\{\mathbf A^{(2)}, \mathbf A^{(3)}\right\}\), and \(\left\{\mathbf A^{(1)}, \mathbf A^{(2)}, \mathbf A^{(3)}\right\}\) are heterogeneous, because their probability matrices will be different. So, unfortunately, the JRDPG cannot handle the hetereogeneity between the random networks of Facebook and Twitter with the random network for Linkedin. To remove this restrictive homogeneity property of the JRDPG, you can turn to the single network model you just learned about: the Independent Erdos Renyi (IER) random network. Next, you will see how the IER random network can be used in conjunction with the COSIE model to allow you to capture this heterogeneity.
4.7.2. Common Subspace Independent Edge (COSIE) Model#
In your example on social networks, notice that the Facebook and Linkedin connections looked relatively similar, but had an important difference: on Linkedin, there was an administrative meeting, and the employees on the administrative team exchanged far more connections than usual among one another. It turns out that, in fact, the random networks \(\mathbf A^{(1)}\) and \(\mathbf A^{(3)}\) which underly the social networks \(A^{(1)}\) and \(A^{(3)}\) were also different, because the probability matrices were different:
fig, axs = plt.subplots(1,3, figsize=(25, 6))
plot_prob_block_annot(P1, ax=axs[0], title="$P^{(1)}$ Facebook Prob. Matrix")
plot_prob_block_annot(P3, ax=axs[1], title="$P^{(3)}$ Linkedin Prob. Matrix")
plot_prob_block_annot(np.abs(P1 - P3), ax=axs[2], annot=True, title="$|P^{(1)} - P^{(3)}|$")
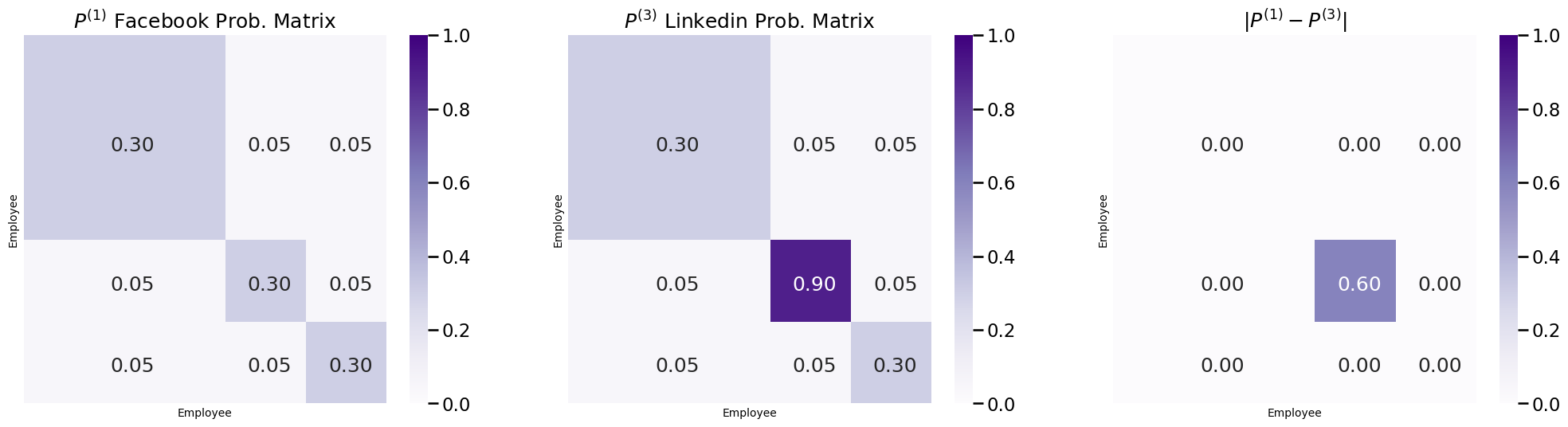
Notice that the difference in the probability of a connection being shared between the members of the administrative team is \(0.6\) higher in the probability matrix for Linkedin than for Facebook. This is because the random networks \(\mathbf A^{(1)}\) and \(\mathbf A^{(3)}\) are heterogeneous. To reflect this heterogeneity, we turn to the COmmon Subspace Independent Edge (COSIE) model.
4.7.4. References#
- 1
Avanti Athreya, Donniell E. Fishkind, Minh Tang, Carey E. Priebe, Youngser Park, Joshua T. Vogelstein, Keith Levin, Vince Lyzinski, and Yichen Qin. Statistical inference on random dot product graphs: a survey. J. Mach. Learn. Res., 18(1):8393–8484, January 2017. doi:10.5555/3122009.3242083.
- 2
Keith Levin, A. Athreya, M. Tang, V. Lyzinski, and C. Priebe. A Central Limit Theorem for an Omnibus Embedding of Multiple Random Dot Product Graphs. 2017 IEEE International Conference on Data Mining Workshops (ICDMW), 2017. URL: https://www.semanticscholar.org/paper/A-Central-Limit-Theorem-for-an-Omnibus-Embedding-of-Levin-Athreya/fdc658ee1a25c511d7da405a9df7b30b613e8dc8.
- 3
Jesús Arroyo, Avanti Athreya, Joshua Cape, Guodong Chen, Carey E. Priebe, and Joshua T. Vogelstein. Inference for Multiple Heterogeneous Networks with a Common Invariant Subspace. Journal of Machine Learning Research, 22(142):1–49, 2021. URL: https://jmlr.org/papers/v22/19-558.html.
- 4
Vince Lyzinski, Donniell E. Fishkind, and Carey E. Priebe. Seeded graph matching for correlated Erdös-Rényi graphs. J. Mach. Learn. Res., 15(1):3513–3540, January 2014. doi:10.5555/2627435.2750357.
- 5
Konstantinos Pantazis, Avanti Athreya, Jesús Arroyo, William N. Frost, Evan S. Hill, and Vince Lyzinski. The Importance of Being Correlated: Implications of Dependence in Joint Spectral Inference across Multiple Networks. Journal of Machine Learning Research, 23(141):1–77, 2022. URL: https://jmlr.org/papers/v23/20-944.html.