Semiparametric latent position test - simulations¶
from pkg.utils import set_warnings
import ast
import datetime
import time
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from graspologic.inference import latent_position_test
from graspologic.plot import heatmap
from graspologic.simulations import rdpg
from joblib import Parallel, delayed
from pkg.io import get_out_dir, savefig
from pkg.plot import set_theme
from scipy.stats import kstest
t0 = time.time()
foldername = "semipar_simulations"
set_theme()
def stashfig(name, **kwargs):
savefig(name, foldername=foldername, **kwargs)
Simulation setup¶
Here we sample from a simple RDPG model:
\(d = 1\)
\(n = 64\)
\(X_i \overset{iid}{\sim} Uniform2D(0.1, 0.7)\), where \(Uniform2D(0.1, 0.7)\) means uniform on a square from [0.1, 0.7] in two dimensions.
\(Y_i \overset{iid}{\sim} Uniform2D(0.1, 0.7)\), independent of \(X\)
If directed, \(P = X Y^T\)
If undirected, \(P = XX^T\)
\(A_1, A_2 \overset{iid}{\sim} Bernoulli(P)\)
Using this model, we run the following simulation under the null distribution:
For \(i\) in \(n_{trials}\):
\(A_1, A_2 \overset{iid}{\sim} Bernoulli(P)\)
\(p\) = \(Semipar(A_1, A_2)\)
Append p-value \(p\) to the distribution of p-values under the null
For running semipar, I use \(n_{bootstraps} = 200\). This is how many samples are used to compute the null distribution to compute the p-value for each trial.
The above simulation is run using ASE or Omni as the embedding method/test statistic.
n_verts = 64
X1 = np.random.uniform(0.15, 0.7, size=(n_verts, 2))
Y1 = np.random.uniform(0.15, 0.7, size=(n_verts, 2))
P = X1 @ Y1.T
print(f"Maximum expected degree: {np.max(P.sum(axis=0))}")
print(f"Minimum expected_degree: {np.min(P.sum(axis=0))}")
A1 = rdpg(X1, Y1, directed=True)
heatmap(A1)
Maximum expected degree: 35.58650928605349
Minimum expected_degree: 10.471348334562961
<AxesSubplot:>

def run_semipar_experiment(
seed=None,
directed=True,
perturbation="none",
n_verts=64,
n_bootstraps=200,
methods=["ase", "omnibus"],
):
np.random.seed(seed)
# sample latent positions
X1 = np.random.uniform(0.15, 0.7, size=(n_verts, 2))
if directed:
Y1 = np.random.uniform(0.15, 0.7, size=(n_verts, 2))
else:
Y1 = None
if perturbation == "none":
X2 = X1
Y2 = Y1
# sample networks
A1 = rdpg(X1, Y1, directed=directed, loops=False, rescale=False)
A2 = rdpg(X2, Y2, directed=directed, loops=False, rescale=False)
# run the tests
rows = []
for method in methods:
lpt_result = latent_position_test(
A1,
A2,
n_components=2,
workers=1,
embedding=method,
n_bootstraps=n_bootstraps,
)
lpt_result = lpt_result._asdict()
lpt_result["method"] = method
lpt_result["perturbation"] = perturbation
lpt_result["directed"] = directed
lpt_result = {**lpt_result, **lpt_result["misc_stats"]}
lpt_result.pop("misc_stats")
lpt_result["null_distribution_1"] = list(lpt_result["null_distribution_1"])
lpt_result["null_distribution_2"] = list(lpt_result["null_distribution_2"])
rows.append(lpt_result)
return rows
out_dir = get_out_dir(foldername=foldername)
def arrayize(string):
return np.array(ast.literal_eval(string), dtype=float)
def saveload(name, results):
if results is not None:
results.to_csv(out_dir / name)
results = pd.read_csv(
out_dir / name,
index_col=0,
na_values="",
keep_default_na=False,
converters=dict(
null_distribution_1=arrayize,
null_distribution_2=arrayize,
),
)
return results
n_trials = 256
RECOMPUTE = False
if RECOMPUTE:
rng = np.random.default_rng(8888)
all_rows = []
seeds = rng.integers(np.iinfo(np.int32).max, size=n_trials)
all_rows += Parallel(n_jobs=-2, verbose=10)(
delayed(run_semipar_experiment)(seed, False) for seed in seeds
)
seeds = rng.integers(np.iinfo(np.int32).max, size=n_trials)
all_rows += Parallel(n_jobs=-2, verbose=10)(
delayed(run_semipar_experiment)(seed, True) for seed in seeds
)
results = []
for rows in all_rows:
results += rows
results = pd.DataFrame(results)
else:
results = None
results = saveload("null-simulation-results", results)
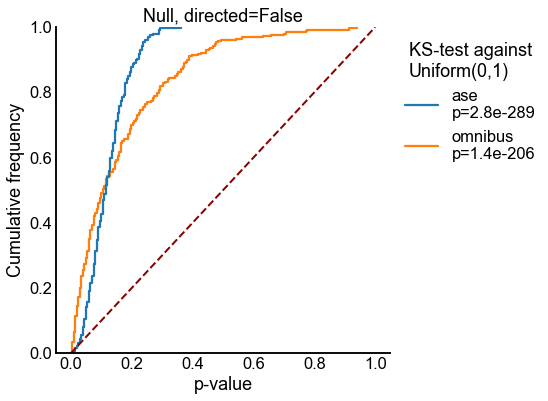
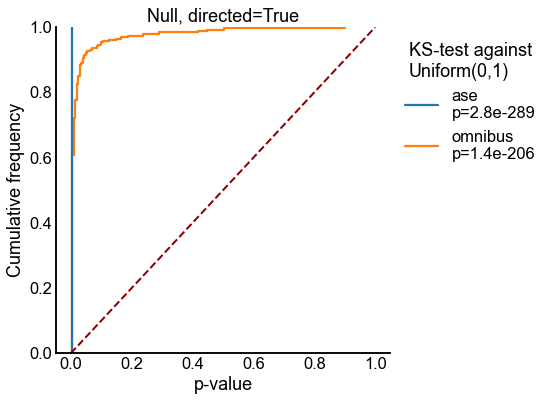
Plot ECDFs of p-values under the null¶
for directed in [False, True]:
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
sns.ecdfplot(
data=results[(results["directed"] == directed)],
x="p_value",
hue="method",
)
ax.plot([0, 1], [0, 1], linestyle="--", linewidth=2, color="darkred")
ax.set(
title=f"Null, directed={directed}",
xlabel="p-value",
ylabel="Cumulative frequency",
)
legend = ax.get_legend()
handles = legend.legendHandles
labels = legend.get_texts()
legend.remove()
labels = [label.get_text() for label in labels]
new_labels = []
for method in labels:
pvalues = results[results["method"] == method]["p_value"]
stat, pvalue = kstest(pvalues, "uniform", args=(0, 1), alternative="greater")
new_labels.append(f"{method}\np={pvalue:0.2g}")
ax.legend(
handles=handles,
labels=new_labels,
bbox_to_anchor=(1, 1),
loc="upper left",
title="KS-test against\nUniform(0,1)",
)
stashfig(f"semipar-null-ecdf-directed={directed}")
End¶
elapsed = time.time() - t0
delta = datetime.timedelta(seconds=elapsed)
print("----")
print(f"Script took {delta}")
print(f"Completed at {datetime.datetime.now()}")
print("----")
----
Script took 0:00:02.491038
Completed at 2021-05-03 09:04:20.007807
----
