Testing bilateral symmetry - semiparametric test¶
Preliminaries¶
import datetime
import pprint
import time
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from giskard.plot import scatterplot
from graspologic.inference import latent_position_test
from graspologic.utils import binarize, multigraph_lcc_intersection, symmetrize
from pkg.data import load_adjacency, load_node_meta
from pkg.io import get_out_dir, savefig
from pkg.plot import set_theme
from pkg.utils import get_paired_inds, get_paired_subgraphs, set_warnings
# from src.visualization import adjplot # TODO fix graspologic version and replace here
set_warnings()
t0 = time.time()
RECOMPUTE = False
foldername = "semipar"
def stashfig(name, **kwargs):
savefig(name, foldername=foldername, **kwargs)
out_dir = get_out_dir(foldername=foldername)
colors = sns.color_palette("Set1")
palette = dict(zip(["Left", "Right"], colors))
set_theme()
/Users/bpedigo/miniconda3/envs/maggot-revamp/lib/python3.8/site-packages/umap/__init__.py:9: UserWarning: Tensorflow not installed; ParametricUMAP will be unavailable
warn("Tensorflow not installed; ParametricUMAP will be unavailable")
Load the data¶
Load node metadata and select the subgraphs of interest¶
meta = load_node_meta()
meta = meta[meta["paper_clustered_neurons"]]
adj = load_adjacency(graph_type="G", nodelist=meta.index)
lp_inds, rp_inds = get_paired_inds(meta)
left_meta = meta.iloc[lp_inds]
right_meta = meta.iloc[rp_inds]
ll_adj, rr_adj, lr_adj, rl_adj = get_paired_subgraphs(adj, lp_inds, rp_inds)
# TODO not sure what we wanna do about LCCs here
adjs, lcc_inds = multigraph_lcc_intersection([ll_adj, rr_adj], return_inds=True)
ll_adj = adjs[0]
rr_adj = adjs[1]
print(f"{len(lcc_inds)} in intersection of largest connected components.")
print(f"Original number of valid pairs: {len(lp_inds)}")
left_meta = left_meta.iloc[lcc_inds]
right_meta = right_meta.iloc[lcc_inds]
meta = pd.concat((left_meta, right_meta))
n_pairs = len(ll_adj)
print(f"Number of pairs after taking LCC intersection: {n_pairs}")
1210 in intersection of largest connected components.
Original number of valid pairs: 1211
Number of pairs after taking LCC intersection: 1210
Run a latent position test¶
if RECOMPUTE:
preprocess = [symmetrize, binarize]
graphs = [ll_adj, rr_adj]
for func in preprocess:
for i, graph in enumerate(graphs):
graphs[i] = func(graph)
ll_adj = graphs[0]
rr_adj = graphs[1]
n_bootstraps = 200
test_case = "rotation"
embedding = "ase"
verbose = 1
workers = -2
rows = []
for embedding in ["ase", "omnibus"]:
for n_components in np.arange(6, 15):
currtime = time.time()
params = dict(
embedding=embedding,
n_components=n_components,
test_case=test_case,
n_bootstraps=n_bootstraps,
workers=workers,
)
pvalue, tstat, misc = latent_position_test(ll_adj, rr_adj, **params)
elapsed = time.time() - currtime
row = params.copy()
row["pvalue"] = pvalue
row["tstat"] = tstat
rows.append(row)
results = pd.DataFrame(rows)
results.to_csv(out_dir / "semipar_results")
if verbose > 0:
pprint.pprint(row)
print()
results = pd.read_csv(out_dir / "semipar_results", index_col=0)
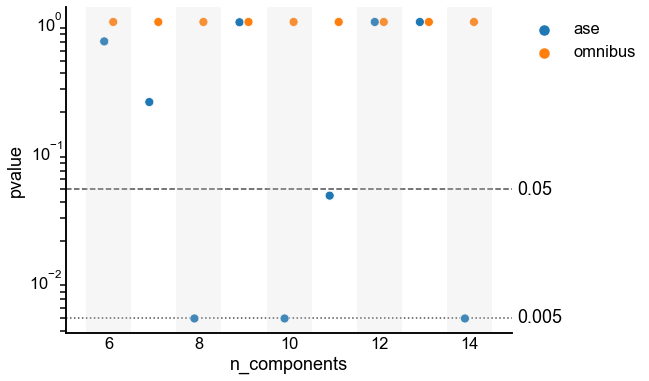
Plot p-values¶
ax = scatterplot(
data=results,
x="n_components",
y="pvalue",
hue="embedding",
shift="embedding",
shade=True,
)
ax.set_yscale("log")
styles = ["--", ":"]
line_locs = [0.05, 0.005]
line_kws = dict(color="black", alpha=0.7, linewidth=1.5, zorder=-1)
for loc, style in zip(line_locs, styles):
ax.axhline(loc, linestyle=style, **line_kws)
ax.text(ax.get_xlim()[-1] + 0.1, loc, loc, ha="left", va="center")
stashfig("semipar-pvalues-by-dimension")
# #%%
# test_case = "rotation"
# embedding = "omnibus"
# n_components = 8
# n_bootstraps = 100
# n_repeats = 5
# rows = []
# for n_shuffle in [4, 8, 16]:
# for repeat in range(n_repeats):
# inds = np.arange(len(rr_adj))
# choice_inds = np.random.choice(len(rr_adj), size=n_shuffle, replace=False)
# shuffle_inds = choice_inds.copy()
# np.random.shuffle(shuffle_inds)
# inds[choice_inds] = inds[shuffle_inds]
# rr_adj_shuffle = rr_adj[np.ix_(inds, inds)]
# currtime = time.time()
# pvalue, tstat, misc = latent_position_test(
# ll_adj,
# rr_adj_shuffle,
# embedding=embedding,
# n_components=n_components,
# test_case=test_case,
# n_bootstraps=n_bootstraps,
# )
# row = {
# "pvalue": pvalue,
# "tstat": tstat,
# "n_shuffle": n_shuffle,
# "n_components": n_components,
# "n_bootstraps": n_bootstraps,
# "embedding": embedding,
# "test_case": test_case,
# "repeat": repeat,
# }
# rows.append(row)
# print(f"{time.time() - currtime:.3f} seconds elapsed.")
# print(f"n_shuffle: {n_shuffle}")
# print(f"test case: {test_case}")
# print(f"embedding: {embedding}")
# print(f"n_components: {n_components}")
# print(f"p-value: {pvalue}")
# print(f"tstat: {tstat}")
# print()
# #%%
# results = pd.DataFrame(rows)
# fig, ax = plt.subplots(1, 1, figsize=(8, 6))
# sns.scatterplot(data=results, x="n_shuffle", y="pvalue", ax=ax)
# stashfig("shuffle-p-values")
End¶
elapsed = time.time() - t0
delta = datetime.timedelta(seconds=elapsed)
print("----")
print(f"Script took {delta}")
print(f"Completed at {datetime.datetime.now()}")
print("----")
----
Script took 0:00:02.892378
Completed at 2021-04-27 12:59:46.906761
----
