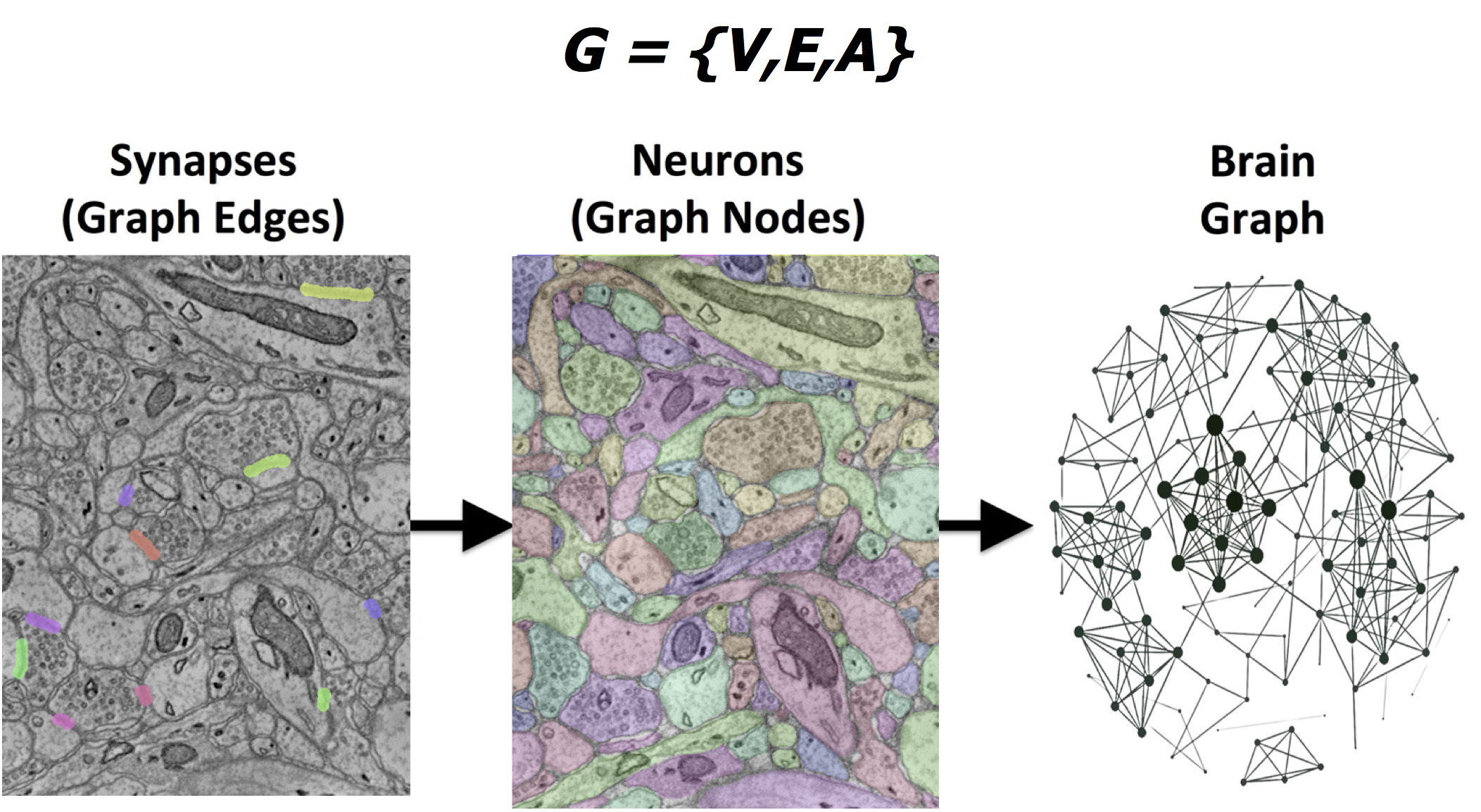
Generating Graphs¶
A graph is composed of vertices (neurons), edges (synapses) and their attributes (e.g., weight, direction). To construct graphs in the RAMON framework, we associate RAMONSynapses with RAMONSegments. RAMONSegments are grouped into neurons.
For automated parsing methods, the synapse-segment links are constructed by determining the two segments with the greatest spatial overlap for each synapse. For manual methods, these links are recorded in a table. Either way, the relationship can be recorded bidirectionally in RAMONSegment.synapse and RAMONSynapse.segment.
Once the object relationships have been stored in the NeuroData database, a graph can be retrieved by efficiently traversing metadata as depicted in the figure below. Typically we start at synapses and work up to segments and neurons. Users should specify parameters for the following function:
edgeList = graph_retrieval(synLocation, synToken, synChannel, synResolution, synIdList, neuLocation, neuToken, neuChannel, neuResolution, segGraph)
AC4 Graph¶
synLocation = 'openconnecto.me';
synToken = 'test_ramonify_public';
synChannel = 'synapse';
synResolution = 1;
synIdList = [];
neuLocation = 'openconnecto.me';
neuToken = 'test_ramonify_public';
neuChannel = 'neuron';
neuResolution = 1;
segGraph = 1;
% Retrieve edge list
eListOut = graph_retrieval(synLocation, synToken, synChannel, synResolution, synIdList, neuLocation, neuToken, neuChannel, neuResolution, segGraph);
% Make graphs
[neuGraph, nId, synGraph, synId] = graph_matrix(eListOut);
Kasthuri Cylinder Graph¶
synLocation = 'openconnecto.me';
synToken = uploadToken;
synChannel = 'synapses';
synResolution = 3;
neuLocation = 'openconnecto.me';
neuToken = uploadToken;
neuChannel = 'neurons';
neuResolution = 3;
edgeList = graph_retrieval(synLocation, synToken, synChannel, synResolution, [], neuLocation, neuToken, neuChannel, neuResolution, 0)
attredgeWriter(edgeList,'kasthuri_graph_v1.attredge')
Post Processing¶
The output of graph_retrieval is an edgeList, which has fields for the ids of two neurons, the corresponding synapse, direction, and synapse correspondence (used for matching with truth). These graphs can be visualized using graph_matrix or saved using the file attredgeWriter, the result of which can be uploaded to MROCP to convert to GraphML or other formats.