vesicle-rf¶
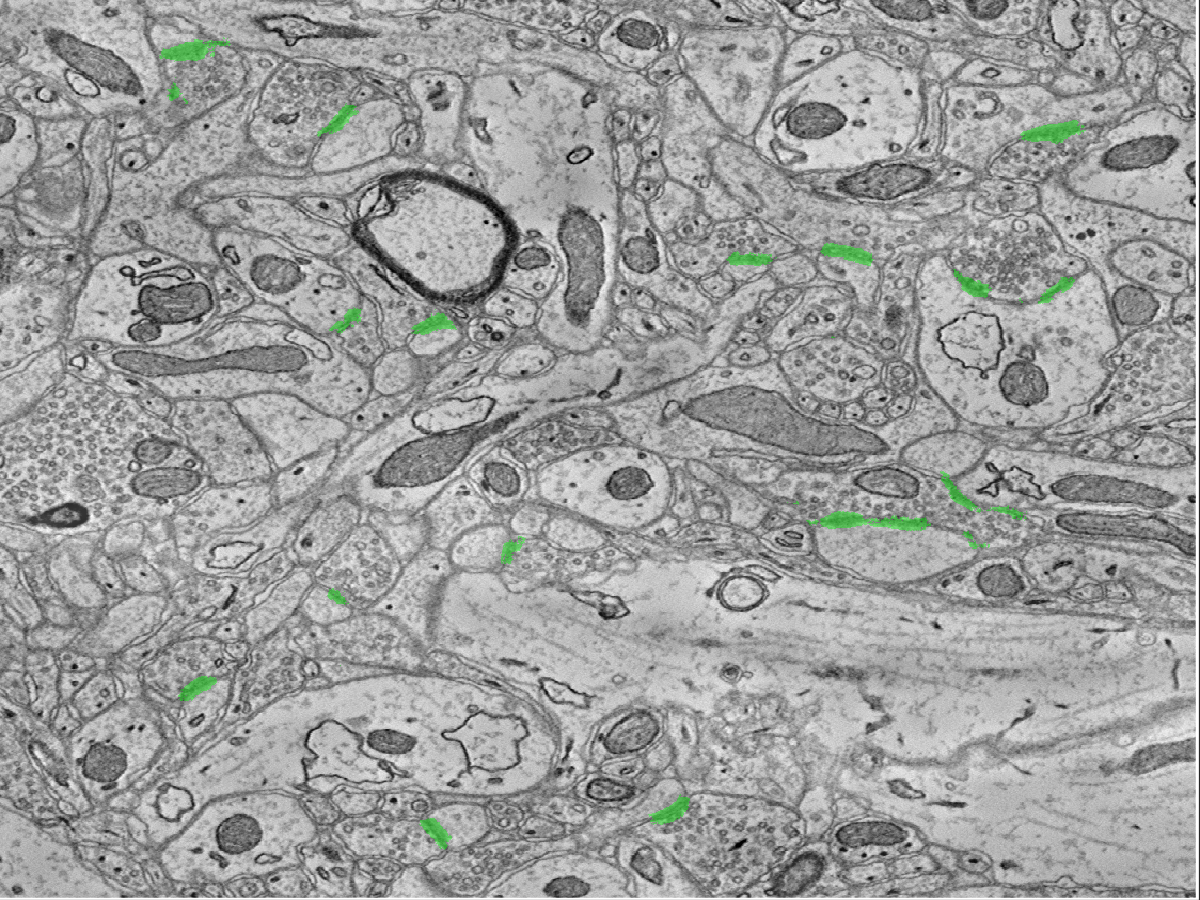
To find synapses (i.e., graph edges), we develop a lightweight, scalable Random Forest-based synapse classifier. We combine texture (i.e., intensity measures, image gradient magnitude, local binary patterns, structure tensors) with biological context (i.e., membranes and vesicles). We pruned a large feature set to just ten features and used this information to train a random forest. The pixel-level probabilities produced by the classifier were grouped into objects using size and probability thresholds and a connected component analysis. This method requires substantially less RAM than previous methods, which enables large-scale processing. A key insight was identifying neurotransmitter-containing vesicles present near (chemical, mammalian) synapses.
These were located using a lightweight template correlation method and clustering. Performance was further enhanced by leveraging the high probability membrane voxels to restrict our search, improving both speed and precision. Our synapse performance was evaluated using the F1 object detection score, computed as the harmonic mean of the precision and recall, based on a gold-standard, neuroanatomist-derived dataset. We took care to define our object detection metric to disallow a single detection counting for multiple true synapses (as that result is ambiguous and allows for a single detection covering the whole volume to produce an F1 score of 1).
vesicle-rf is divided into three major sections: training, evaluation, and deployment.
Validation (Quick Start)¶
To get a sense of how the classifier works, we recommend first running the following function. It leverages a pre-trained classifier and evaluates performance on a small test volume.
Performance metrics are imperfect (due to small volume issues), but you should achieve results similar to the following:
metrics =
precision: 0.8966
recall: 0.6047
TP: 26
FP: 3
FN: 17
% function vesiclerf_example_short()
% Driver function to demonstrate vesicle-rf functionality for new users.
% This is an abbreviated version in the interest of time.
%
% **Inputs**
%
% None. Driver script is self-contained.
%
% **Outputs**
%
% None. Driver script is self-contained.
%
% **Notes**
%
% Using the default parameters, small edge synapses may be missed in this reference implementation.
% As a prerequisite to running this script, you should have a trained classifier (provided in the git repo)
% called bmvc_classifier - you can use a different classifier if desired
if nargin < 2
zStart = 1000;
zStop = 1025;
end
zSlice = [zStart, zStop];
padX = 50; padY = 50; padZ = 2;
%Get data
[eData, mData, sData, vData] = get_ac3_data(zSlice(1),zSlice(2), padX, padY, padZ);
% Save Data in an appropriate format
cube = eData.clone;
save('eData.mat','cube');
cube = mData.clone;
save('mData.mat','cube');
cube = sData.clone;
save('sData.mat','cube');
cube.setCutout(cube.data(1+padX:end-padX, 1+padY:end-padY, 1+padZ:end-padZ));
save('sDataPR.mat','cube');
cube = vData.clone;
save('vData.mat','cube');
% When deploying vesicle-rf, the core of the code consists of
% vesiclerf_probs and vesiclerf_object
%% PIXEL CLASSIFICATION
vesiclerf_probs('edata', 'vData', 'mData', 'bmvc_classifier', padX, padY, padZ, 'classProbTest.mat')
%% OBJECT PROCESSING
vesiclerf_object('classProbTest.mat', 0.90, 0, 5000, 2000, 1, 0, 0, 'testObjVol.mat', 0, 0, 0)
%% UPLOAD
if 0 % sample upload - please provide your own token and channels
server = 'openconnecto.me';
token = 'vesicle_example';
channel = 'prob';
cubeUploadDense(server, token, channel, 'testObjVol', 'RAMONSynapse', 0)
channel = 'object'
end
%% METRICS COMPUTATION
pr_objects('testObjVol','sDataPR','metrics_short')
metrics_short
% visualize results
load eData
eData = cube;
load testObjVol
h = image(cube); h.associate(cube)
Preprocessing¶
Membrane detection is accomplished using caffe and is documented separately. vesicle-rf assumes that membrane detection is completed for the region of interest prior to synapse detection.
Vesicle detection (here referring to neurotransmitter-containing vesicles) is accomplished using template matching (derived from the source dataset).
Vesicle identification may be run using the vesicle_detect_quick.m function, which is documented in Functions.
Training¶
A training workflow exists to download manual annotations and data and build a classifier, using the vesiclerf_train.m function. This function uses a combination of raw image data, manual truth labels, and computer vision results to train a classifier for downstream use.
Evaluation¶
Once a classifier has been trained, a test volume can be evaluated using the functions: vesiclerf_probs to compute pixel-level synapse probabilities, and vesiclerf_object.m, which post-processes the data to identify putative synaptic objects. pr_evaluate_full is an evaluation function designed to grid search over the range of possible parameters to construct a precision-recall curve.
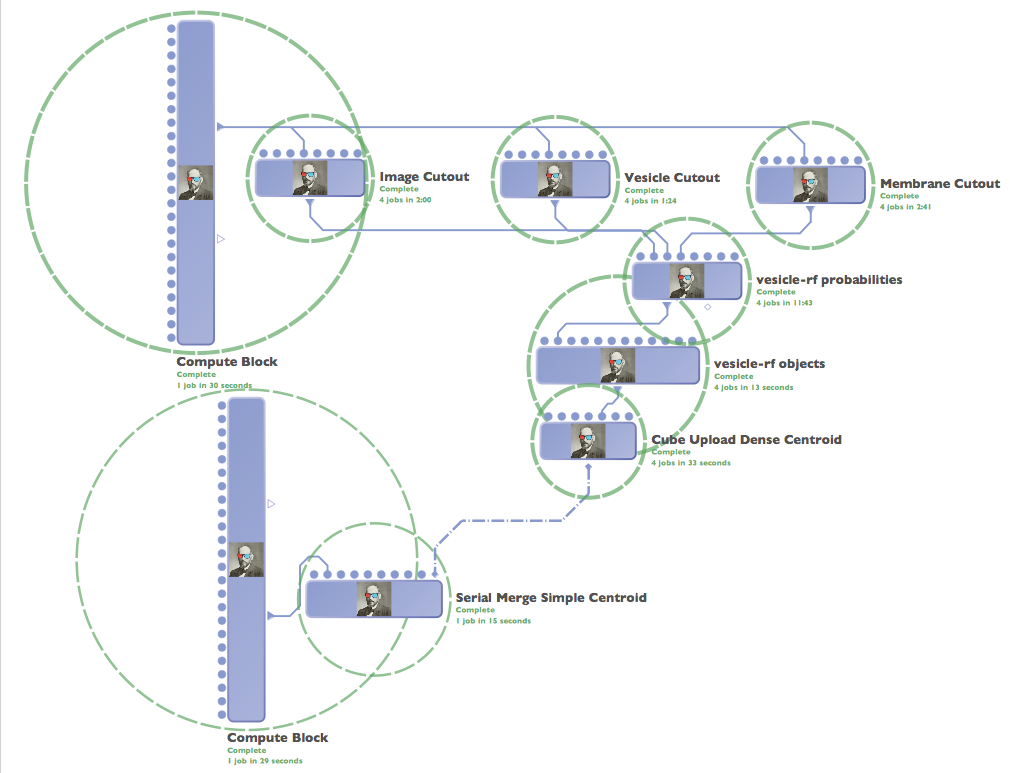
Deployment¶
A deployment workflow - based on an operating point chosen in the evaluation step runs in LONI and includes functionality to merge across cuboid boundaries and compute centroids for each object.
Example Usage¶
We recommend learning to use our classifier by using the quick start above. You may train and evaluate your own classifier by running two functions. These take a total of about 2 hours to complete, due largely to the inefficient and large parameter space swept after classification.
Running the full string can be accomplished by executing:
vesicle_rf_train('your_classifier')
vesicle_rf_example() %change the script to reflect your classifier, if needed.