# Kasthuri 2015 Cell Paper Data #
The raw data used to generate the analyses in the [Kasthuri 2015 Cell Paper](http://www.cell.com/abstract/S0092-8674(15)00824-7) can be downloaded via the Open Connectome Project. Datasets can be visualized using [CATMAID](http://openconnecto.me/catmaid/?dataview=13).
The following tokens are considered to be authoritative, as of 08.11.2015. Tokens in brackets are the raw, unadulterated version of the data, read only and available to the public.
**Original Electron Microscopy data**
* kasthuri11: http://openconnecto.me/ocp/ca/kasthuri11/info/
**Color Corrected Electron Microscopy data**
* kasthuri11cc: http://openconnecto.me/ocp/ca/kasthuri11cc/info/
**Low Resolution dataset**
* kasthuri14Maine: http://openconnecto.me/ocp/ca/kasthuri14Maine/info/
**Medium Resolution dataset and annotations**
* kasthuri14s1colEM: http://openconnecto.me/ocp/ca/kasthuri14s1colEM/info/
* kasthuri14s1colANNO: http://openconnecto.me/ocp/ca/kasthuri14s1colANNO/info/
**Small High Resolution Annotated Regions of Interest**
* ac3: http://openconnecto.me/ocp/ca/ac3/info/
* ac4: http://openconnecto.me/ocp/ca/ac4/info/
**High Resolution Annotated Data: neuron segments, synapses, vesicles, and mitochondria have been annotated at Scale 1 (6 x 6 x 30 nm).**
* kat11segments: http://openconnecto.me/ocp/ca/kat11segments/info/
* kat11synapses: http://openconnecto.me/ocp/ca/kat11synapses/info/
* kat11vesicles: http://openconnecto.me/ocp/ca/kat11vesicles/info/
* kat11mito: http://openconnecto.me/ocp/ca/kat11mito/info/
**Cylinders defining core annotation regions**
* kat11mojocylinder: http://openconnecto.me/ocp/ca/kat11mojocylinder/info/
* kat11redcylinder: http://openconnecto.me/ocp/ca/kat11redcylinder/info/
* kat11greencylinder: http://openconnecto.me/ocp/ca/kat11greencylinder/info/
### Data Access ###
Open Connectome Project provides a variety of methods to access the data described above. Data access is by RESTful call, and is explained here: openconnectome.github.io/open-connectome. CAJAL provides a MATLAB interface, and OCPy provides a beta python package for accessing data.
We strongly recommend downloading data in chunks for processing and analysis, using our spatial database to make research easier. However, if you prefer flat files (PNGs), you may download them directly using the following links. If you do decide to download the entire dataset, please download the tar files below for faster access and to reduce server load - total download is hundreds of gigabytes.
A simple example of a RESTful call which downloads 50MB of the orginal dataset is:
`http://openconnecto.me/ocp/ca/kasthuri11cc/hdf5/1/3000,4000/5000,6000/1000,1050/`
The first slice can be viewed here:
`http://openconnecto.me/ocp/ca/kasthuri11cc/xy/1/3000,4000/5000,6000/1000/`
### Data Download Examples ###
#### Using CAJAL ####
CAJAL is a MATLAB toolbox that implements the RAMON standard and provides simple functionality to get and put data to OCP. It can be downloaded here: www.github.com/openconnectome/cajal
The example below downloads raw image and truth labels (annotation data) for 100MB of data, for a region known as AC4.
```
%% Download Data Example
% View data cuboid here (100 megavoxels)
% http://openconnecto.me/ocp/overlay/0.7/ac4/xy/1/4400,5424/5440,6464/1100/
% Download and install CAJAL (www.github.com/openconnectome/cajal)
oo = OCP();
oo.setServerLocation('http://openconnecto.me');
oo.setImageToken('kasthuri11cc');
oo.setAnnoToken('ac4')
q = OCPQuery;
q.setType(eOCPQueryType.annoDense);
q.setCutoutArgs([4400,5424],[5440,6464],[1100,1200],1);
anno = oo.query(q);
q = OCPQuery;
q.setType(eOCPQueryType.imageDense);
q.setCutoutArgs([4400,5424],[5440,6464],[1100,1200],1);
im = oo.query(q);
% Visualize results
h = image(im); h.associate(anno);
```
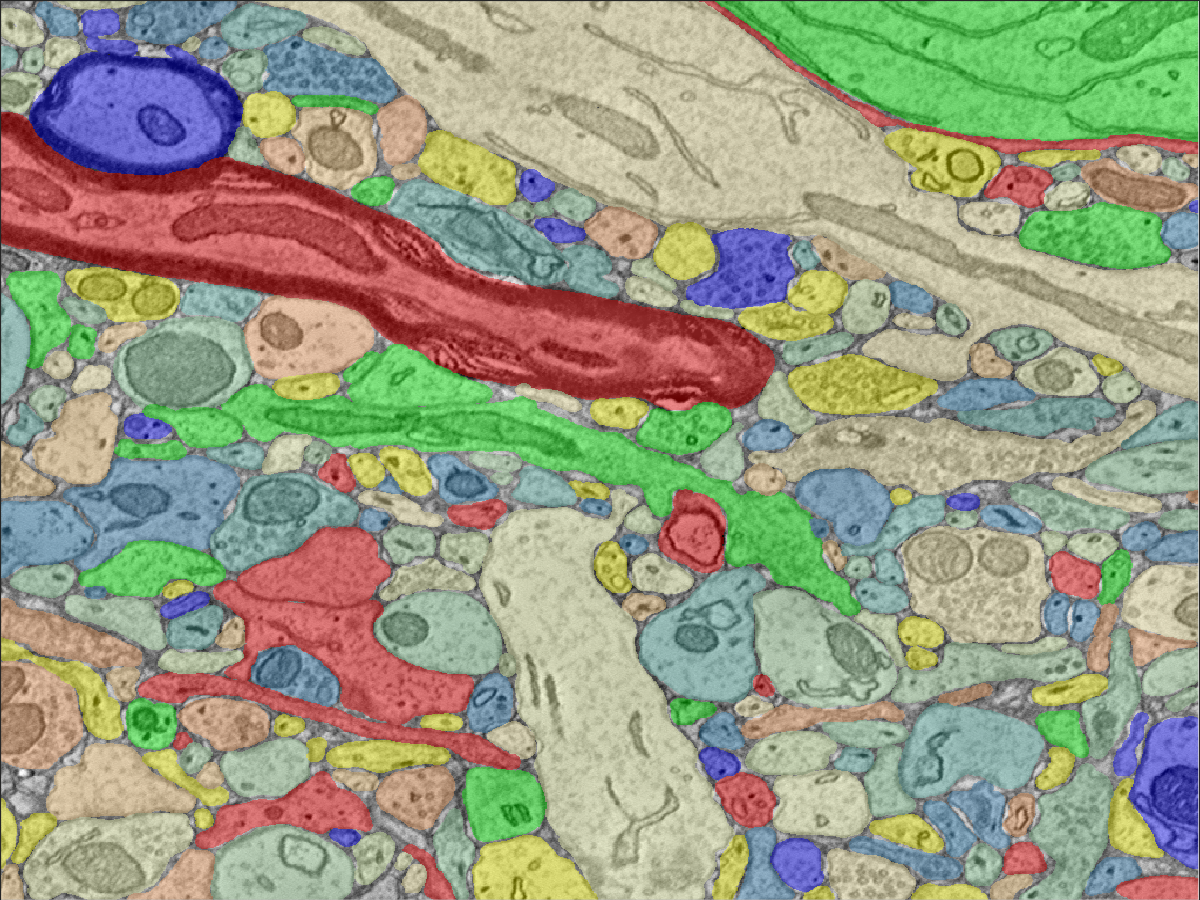
#### Using OCPy ####
We also provide a [Python package](https://github.com/openconnectome/ndio) that makes downloading raw data easier. To install, simply use pip:
pip install ndio
If you cannot use pip, then simply clone the git repository using the following command on a unix or OSX operating system:
git clone https://github.com/openconnectome/ndio ndio
cd ndio/packages
You now have access to the **ocpy** package. We can use the below python code to get information about and download the **Kasthuri15** dataset.
After this completes, you'll be left with two folders: **hdf5**, holding 16-slice HDF5 files, and **png/** which will hold one PNG image per z-layer of your data.
#### Prepared Data Downloads ####
Rather than using the Python script above, or CAJAL, you can download full datasets at the following location. Each tar.gz file contains the full dataset for that token, with each z-slice saved as a PNG file. The most commonly requested datasets can be found here: [Kasthuri2015 Data Download](http://openconnecto.me/data/public/kasthuri2015)
Initially, this directory includes the raw EM data (kasthuri11), as well as kat11synapses, kat11segments, kat11vesicles, and kat11mito. If you have any further questions about this data, please post them [here](https://github.com/openconnectome/kasthuri2015/issues).
### RAMON Objects ###
Objects with metadata to facilitate scientific analysis are coming soon!
 #### Using OCPy ####
We also provide a [Python package](https://github.com/openconnectome/ndio) that makes downloading raw data easier. To install, simply use pip:
pip install ndio
If you cannot use pip, then simply clone the git repository using the following command on a unix or OSX operating system:
git clone https://github.com/openconnectome/ndio ndio
cd ndio/packages
You now have access to the **ocpy** package. We can use the below python code to get information about and download the **Kasthuri15** dataset.
After this completes, you'll be left with two folders: **hdf5**, holding 16-slice HDF5 files, and **png/** which will hold one PNG image per z-layer of your data.
#### Prepared Data Downloads ####
Rather than using the Python script above, or CAJAL, you can download full datasets at the following location. Each tar.gz file contains the full dataset for that token, with each z-slice saved as a PNG file. The most commonly requested datasets can be found here: [Kasthuri2015 Data Download](http://openconnecto.me/data/public/kasthuri2015)
Initially, this directory includes the raw EM data (kasthuri11), as well as kat11synapses, kat11segments, kat11vesicles, and kat11mito. If you have any further questions about this data, please post them [here](https://github.com/openconnectome/kasthuri2015/issues).
### RAMON Objects ###
Objects with metadata to facilitate scientific analysis are coming soon!
#### Using OCPy ####
We also provide a [Python package](https://github.com/openconnectome/ndio) that makes downloading raw data easier. To install, simply use pip:
pip install ndio
If you cannot use pip, then simply clone the git repository using the following command on a unix or OSX operating system:
git clone https://github.com/openconnectome/ndio ndio
cd ndio/packages
You now have access to the **ocpy** package. We can use the below python code to get information about and download the **Kasthuri15** dataset.
After this completes, you'll be left with two folders: **hdf5**, holding 16-slice HDF5 files, and **png/** which will hold one PNG image per z-layer of your data.
#### Prepared Data Downloads ####
Rather than using the Python script above, or CAJAL, you can download full datasets at the following location. Each tar.gz file contains the full dataset for that token, with each z-slice saved as a PNG file. The most commonly requested datasets can be found here: [Kasthuri2015 Data Download](http://openconnecto.me/data/public/kasthuri2015)
Initially, this directory includes the raw EM data (kasthuri11), as well as kat11synapses, kat11segments, kat11vesicles, and kat11mito. If you have any further questions about this data, please post them [here](https://github.com/openconnectome/kasthuri2015/issues).
### RAMON Objects ###
Objects with metadata to facilitate scientific analysis are coming soon!